MBX v1.2: Accelerating data-driven many-body molecular dynamics simulations.
S. Gupta, E.F. Bull-Vulpe, H. Agnew, S. Iyer, X. Zhu, R. Zhou, C. Knight, F. Paesani, J. Chem. Theory Comput. 21, 1838 (2025).
https://doi.org/10.1021/acs.jctc.4c01333
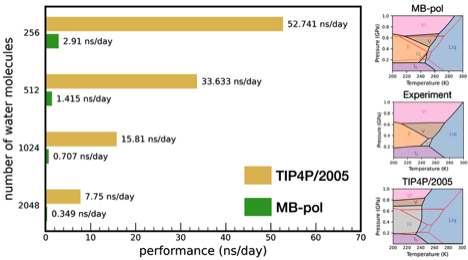
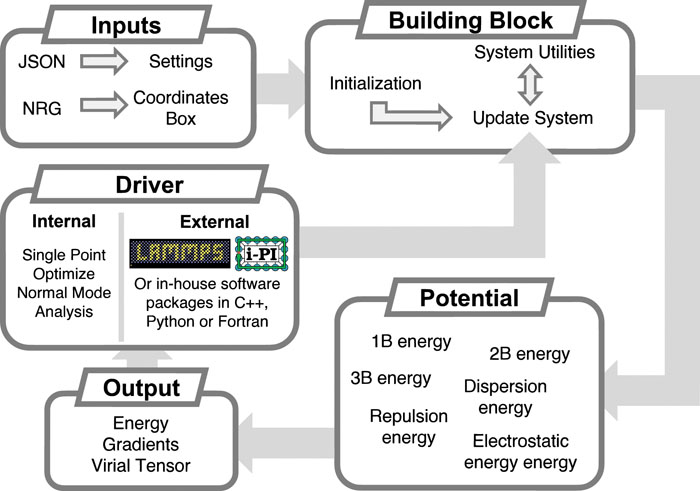
The MBX software provides an advanced platform for molecular dynamics simulations, leveraging state-of-the-art MB-pol and MB-nrg data-driven many-body potential energy functions. Developed over the past decade, these potential energy functions integrate physics-based and machine-learned many-body terms trained on electronic structure data calculated at the “gold standard” coupled-cluster level of theory. Recent advancements in MBX have focused on optimizing its performance, resulting in the release of MBX v1.2. While the inherently many-body nature of MB-pol and MB-nrg ensures high accuracy, it poses computational challenges. MBX v1.2 addresses these challenges with significant performance improvements, including enhanced parallelism that fully harnesses the power of modern multicore CPUs. These advancements enable simulations on nanosecond time scales for condensed-phase systems, significantly expanding the scope of high-accuracy, predictive simulations of complex molecular systems powered by data-driven many-body potential energy functions.